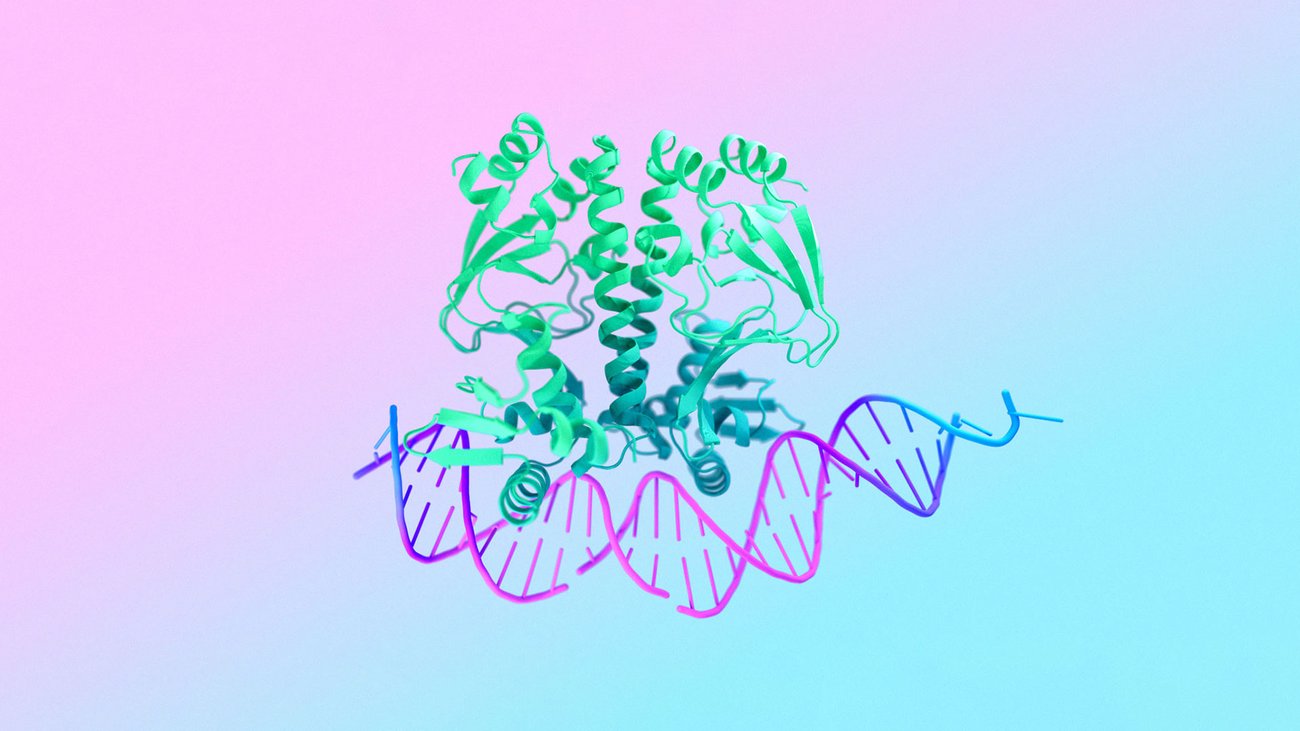
Inside every plant, animal and human cell are billions of molecular machines. They’re made up of proteins, DNA and other molecules, but no single piece works on its own. Only by seeing how they interact together, across millions of types of combinations, can we start to truly understand life’s processes.
In a paper published in Nature, we introduce AlphaFold 3, a revolutionary model that can predict the structure and interactions of all life’s molecules with unprecedented accuracy. For the interactions of proteins with other molecule types we see at least a 50% improvement compared with existing prediction methods, and for some important categories of interaction we have doubled prediction accuracy.
We hope AlphaFold 3 will help transform our understanding of the biological world and drug discovery. Scientists can access the majority of its capabilities, for free, through our newly launched AlphaFold Server, an easy-to-use research tool. To build on AlphaFold 3’s potential for drug design, Isomorphic Labs is already collaborating with pharmaceutical companies to apply it to real-world drug design challenges and, ultimately, develop new life-changing treatments for patients.
Our new model builds on the foundations of AlphaFold 2, which in 2020 made a fundamental breakthrough in protein structure prediction. So far, millions of researchers globally have used AlphaFold 2 to make discoveries in areas including malaria vaccines, cancer treatments and enzyme design. AlphaFold has been cited more than 20,000 times and its scientific impact recognized through many prizes, most recently the Breakthrough Prize in Life Sciences. AlphaFold 3 takes us beyond proteins to a broad spectrum of biomolecules. This leap could unlock more transformative science, from developing biorenewable materials and more resilient crops, to accelerating drug design and genomics research.